- Productos
- Oncohematología
- Inmunología
- Anticuerpos
- Reactivos individuales
- Determinación de compatibilidad cruzada (FCXM)
- FcεR1
- Subclases Ig
- Reactivos individuales
- Kits
- Solicitar información
- Recursos y soporte
- Anticuerpos
- Exosomas
- Reactivos accesorios
- Software
- Tienda
- Servicios
- Síntesis de Péptidos
- Diseño
- Modificación
- Servicios de Proteínas
- Expresión y purificación
- Liofilización
- Desarrollo de anticuerpos
- Monoclonal
- Policlonal
- Servicios especializados de anticuerpos
- OEM/Producción a granel
- Purificación
- Conjugación
- Servicios de exosomas
- Aislamiento y purificación
- Caracterización
- Síntesis de Péptidos
- Soporte
- Sobre nosotros
- Contacto
The role of exosomes in cancer – Part 3
We closed our last blog article talking about phenotyping of exosomes and its importancy in the determination of cellular and subcellular origin, as this provides clues about the characterization of the different exosomal subpopulations, as well as their functionality and the identification of biomarkers related to the disease. Let’s continue there.
▌Bead arrays for exosome phenotyping
Use of Exosome Arrays against tumor proteins has been described, with the aim of phenotyping the population of exosomes. Arrays are powerful tools for antigens study in samples such as plasma, having an advantage over other methods by being able to detect multiple markers simultaneously within a single experiment. However, its introduction in the clinic as a diagnostic tool is limited, beyond the arrays of nucleic acids (DNA and RNA).
Additionally, the specific needs of researchers for multiplex detection of exosomes are heterogeneous, although they can be grouped into four main categories that follow a logical sequence of research and development: surveys of changes in the abundance of exosomes; role in the pathways and physiological and disease states; validation of exosomic biomarkers and clinical diagnosis. This type of experiments generally involve the analysis of samples at different times, stages of disease and types or intensities of treatment, in all patients in a clinical trial and their subsequent comparison, for which technology such as mass spectrometry, are not entirely adequate. In this sense, the Multiplex bead-based Flow Cytometry Assays or bead-based arrays involve attachment of the capture antibody for protein assays to microspheres, and their detection by flow cytometry, are an alternative for multiplex analysis at the clinical level, as demonstrated by the fact of the existence of different platforms of analysis availables from Becton, Dickinson and Company (Cytometric Bead Array) and the partners of Luminex Corp (xMAP™), with a good clinical implantation and with products commercialized for IVD.
THE METHOD
It is a sandwich immunoassay consisting of beads that differ in size or fluorescence intensity, giving rise to color-coded beads, conjugated with capture antibodies that are used for multiplexed immunoassays that differ from each other by their light scatter characteristics. Some of the interesting aspects of bead-based arrays are the fluid phase kinetics, which is faster than the solid phase kinetics of flat matrix; greater accuracy, due to the measurements of hundreds of beads for each analyte; and an easy customisation. Multiplex immunoassays offer a number of advantages over monoplex assays, but for these assays, to be robust they require addressing several questions about dynamic range, cross-reactivity and biological matrix during optimization. For multiplex immunoassays, the effective biological range of each analyte should be considered to ensure that the fluorescence of the detector will fall within the dynamic range of its assay. In the case of monoplex assays, this is approached by a dilution of the sample, but in the multiplex assays analytes with very different concentration ranges can coexist in the same assay, from low concentrations that require high affinity antibodies and at low concentration for coating, or analytes with high concentrations that require a weaker detector or low affinity capture antibodies.
As it is evidenced, antibodies are the molecules on which every test pivots, consequently, the entire system relies on the specificity and sensitivity of the applied antibodies. As for all capture sandwich immunoassays, it is important to confirm that the pair of antibodies used recognizes different epitopes. On the other hand, an additional difficulty in multiplex assays is the cross-reactivity, which arises because the detection antibodies are mixed. This problem increase exponentially with the number of markers and usually requires optimization. A list of exosomic identified markers is provided, where it is depicted the identified markers by means of protein arrays (including the info of the particular antibodies employed for the systematic detection). This is the table that we already included in our last blog article:
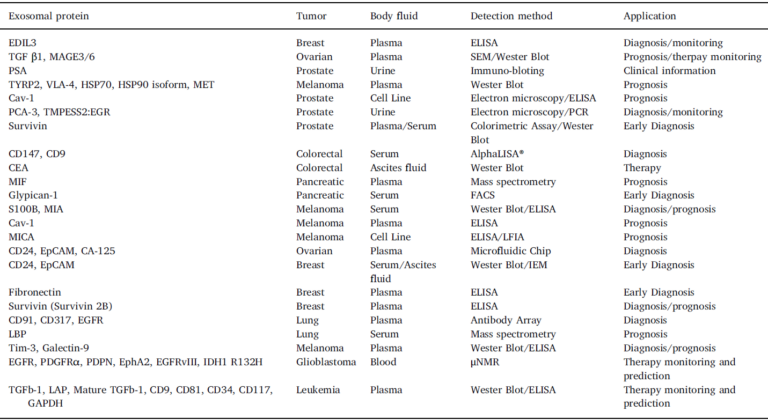
IMPROVING THE RESULTS
In this way, the optimization of the multiplex tests, goes through the choice of the blocking and dilution agents of sample, volume of reaction, number of microspheres per reaction, concentration of capture reagent for coating, detector antibody concentration, washing steps, incubation times and sample volume, although for this last parameter it is not always an optimizable variable, since in many cases the multiplexed array immunoassays are tests under conditions termed as analysis of environmental analyte, allowing measurements where the amount of analyte captured from the solution would directly reflect its concentration in the test sample. In these ambient analyte conditions, the test is independent of the actual sample volume used and this could also occur in the case of exosomes given their abundance in biological fluids.
In the specific case of exosome multiplex beads based assays, antibody coated beads would be responsible for capturing the different tumor exosomes subpopulations, which could be accurately detected by a cocktail of fluorescently conjugated CD9, CD63 and CD81 antibodies (see figure below), since there is an absence of invariant housekeeping markers which makes comparative analysis difficult, and including these three markers is to ensure detection.
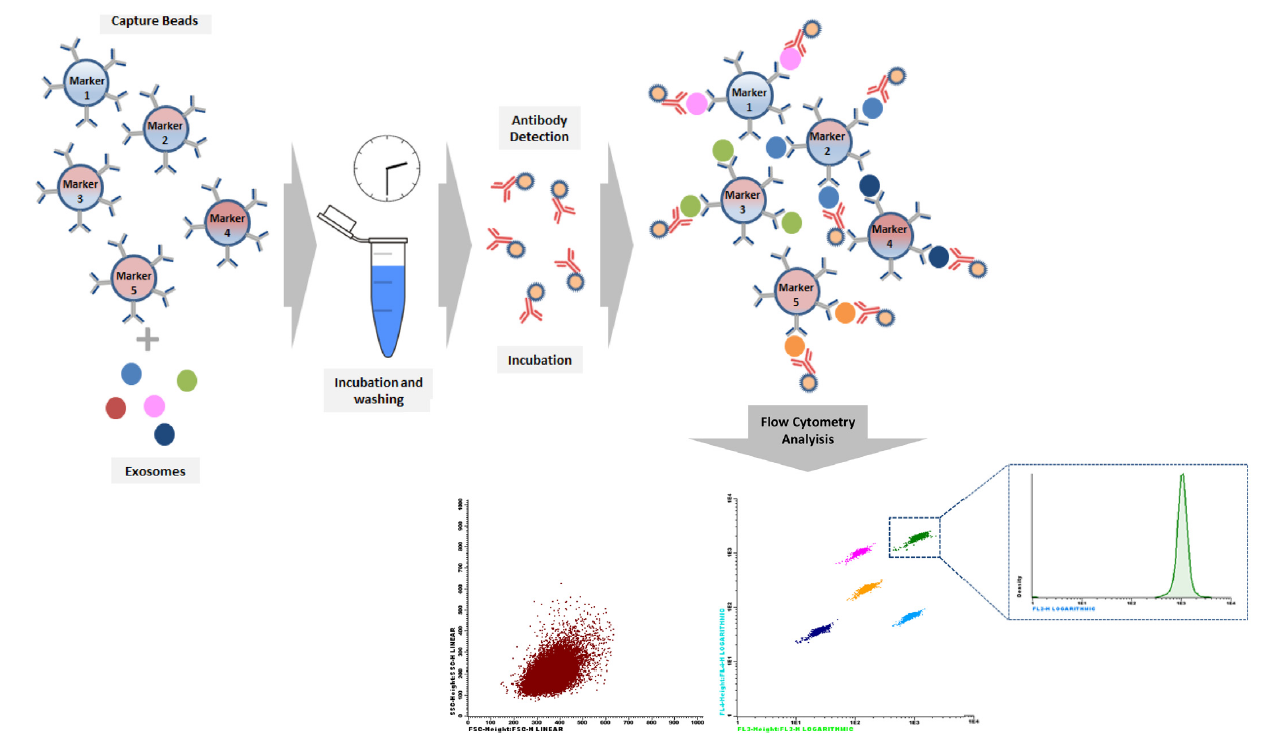
Fig.: Bead-based Array Principle. The arrays utilizes the same basic principle of Monoplex, but in this case each bead set is coated with a specific antibody on the surface and serves as the capture bead for that specific exosome. When a selected panel of antibody capture beads are mixed together and incubated with an unknown sample containing target exosomes, each exosome will be bound by its specific capture bead. After washing, detection pan-exosome antibodies are added and each detection antibody will bind to exosome common markers e.g. CD63, CD9 or CD81, thus forming capture bead-exosome-detection antibody sandwiches and subsequently providing fluorescent signal with different intensities according to the amount of bound exosome. Bead populations come differing levels of APC (FL4) and PerCP (FL3) fluorescence, allowing them to be clearly distinguished from each other. Detection antibodies fluorescently conjugated, usually in PE (FL2), because they are the most sensitive channel, and for each bead population, the PE signal fluorescence intensity is then quantified using a flow cytometer.
ADVANTAGES
Depending on markers selection, bead exosome array would allow (1) to evaluate different subpopulations of exosomes in a single sample; (2) directly phenotype comparison of exosomal subpopulations in different samples; (3) reproducibility and comparative results with previous experiments; (4) a faster evaluation of multiple samples with a single method, discriminating changes in protein composition versus exosome subpopulation; and (5) use of a small amount of sample, which maximizes the amount of exosomes that can be analyzed.
From a current perspective and according to what has been described before in this review, it is not difficult to guess that in a near future it will possible to define and standardize combinations of tumor exosomes specific markers, allowing for diagnosis, prognosis and classification of tumor pathologies, as well as, evaluation of treatment effectiveness. Furthermore if possible, combination of several markers will improve the clinical discrimination between patients with and without cancer.
▌Conclusions and perspectives
There has been an important advance in the molecular characterization of circulating tumor-derived exosomes which have been recognized as a promising source of biomarkers for cancer diagnosis by less invasive procedures such as liquid biopsy.
However, the clinical use of exosomes has been limited by the lack of adequate methods for sensitive detection of exosomes on body fluids and by the absence of specific markers of the different subpopulations of exosomes present in patient samples. In fact, consulting the exocarta, vesiclepedia and EVpedia databases, it can be verified how the most common methods for the identification of most of the markers described have been Western Blotting and Mass Spectrometry. Proteomic techniques that are excellent but difficult to transfer to clinical routine. Provided this, makes sense the development of diagnostic tools based on exosomes features which involves the identification of specific marker profiles of subpopulations of tumor exosomes that are informative of the patient’s condition in terms of diagnosis, prognosis or risk, as is already the case with exosomal microRNA. For its implementation these discoveries have to be accompanied by the development of sensitive, reproducible and easily interpretable methods for the detection and quantification of said subpopulations of exosomes.
Additionally, these methods should be easily implemented in the clinical laboratory, which includes that the method can be carried out in parallel with the rest of clinical analyses on the same type of sample. On this matter, there is a large number of markers that have already been tested in antibody arrays and bead-based array through flow cytometry and that could play an important role in exosome liquid biopsy analysis, due to its high throughput possibilities, its reproducibility, its multiplex capacity and its broad clinical implementation.
The future direction of the exosome bead-based arrays involve the development of specific panels per tumor, which can play an important role in the early diagnosis of disease, in non-invasive cancer monitoring, in the minimal residual disease diagnostic, in the early detection of metastasis, in tumor progression and in drug resistance. Additionally, the development of novel methods to increase the sensitivity of the arrays, allowing the detection of exosomes directly in the samples without the need to isolate or purify them, would greatly contribute to popularize their use. A better sensitivity would also allow the detection of markers with a low expression and the detection of minority subpopulations of exosomes. Likewise, the development of protocols for the quantification of exosomes on the bead arrays, thanks to the use of well-calibrated standards allowing the fluorescence signal to be transferred to a number of particles, also definitively contributed to simplifying the current methods of characterization of exosomes and to popularize this type of arrays.
▌ Immunostep products related with this article:
- Lyophilized Exosome Standards: https://immunostep.com/exosomes/lyophilized-exosome-standards/
- Assay Custom products: https://immunostep.com/custom-services/custom-exosome-services/
- Detection and Characterization of Exosomes: https://immunostep.com/exosomes/exostep-platform-detection-and-characterization/
▌ References:
- Jara-Acevedo, R., Campos-Silva, C., Valés-Gómez, M., Yáñez-Mó, M., Suárez, H., & Fuentes, M. (2019). Exosome beads array for multiplexed phenotyping in cancer. Journal of proteomics, 198, 87–97. https://doi.org/10.1016/j.jprot.2018.12.023